LEARNING OBJECTIVES
In this article, author has explained the primary structure of proteins, peptide bond and determination of primary structure.
The linear sequence of amino acids forming the polypeptides is known as the primary structure of proteins. The polypeptide containing more than 50 amino acids is termed “protein”. Each protein has a unique sequence of amino acids. This sequence is determined by the genes of DNA. The sequence of amino acids determines its function. The change in the sequence of amino acids can cause many genetic disorders.
The amino acid composition of a protein determines its physical and chemical properties.
Peptide bond
Table of Contents
The amino acids are attached by covalent peptide bonds.
Formation of peptide bond
The peptide bond is formed between the amino group of one amino acid and the carboxyl group of the next amino acid with the elimination of the water molecule.
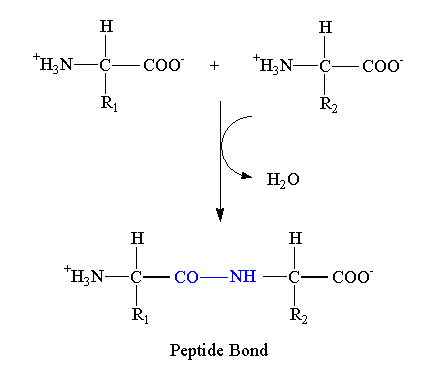
A dipeptide has two amino acids and one peptide bond. Peptides containing more than 10 amino acids are known as polypeptides.
Characteristics of peptide bond
- The peptide bond is rigid and planar.
- It has a partial double bond character.
- It exists in trans configuration.
- Both groups (CO and NH) are polar in nature.
- These groups are involved in hydrogen bond formation.
Writing of peptide structures
The peptide chain is written with a free amino group on the left side (N-terminal residue) and a free carboxyl group on the right side (C-terminal residue). The amino acid sequence is read from N-terminal to C-terminal. The protein biosynthesis also starts from N-terminal.
Representation of peptides
The amino acids in peptides or proteins are represented by three letters or by one letter abbreviation. This is a chemical shorthand to write proteins. For example, glycine is represented by “Gly”, alanine is represented by “Ala”, etc.
Naming of peptides
The peptides are named by removing suffixes like one from glycine, a from tryptophan, ate from glutamate and adding yl except for C-terminal amino acids. Thus, tripeptide composed of glutamate, cysteine, and glycine will name glutamyl-cysteinyl-glycine. Glycine is a C-terminal amino acid.
Determination of the primary structure
The determination of primary structure depends upon the identification of amino acid quality, quantity, and sequence in a protein structure. For the determination of structure, the protein sample should be pure. The determination of primary structure involves three steps;
1. Determination of amino acids composition in protein:
The protein or polypeptide is completely hydrolyzed to release amino acids. These amino acids are determined quantitatively. The hydrolysis is carried by either or base. Enzymes may also use for hydrolysis. Enzyme hydrolysis yields smaller peptides instead of amino acids.
Pronase is a mixture of proteolytic enzymes that cause the complete hydrolysis of proteins.
Separation and estimation of amino acids:
The amino acids released by the hydrolysis of proteins can be determined by chromatography techniques.
2. Degradation of proteins into small fragments
Proteins are composed of polypeptide chains. Separation of the polypeptide chain is essential before degradation
Liberation of polypeptides
When protein is treated with urea or guanidine hydrochloride, it disrupts the non-covalent bonds of protein and breaks the protein into polypeptides. The disulfide linkages are cleaved by treating polypeptide with performic acid.
Number of polypeptides
When protein is treated with dansyl chloride, it determined the number of polypeptides. Dansyl chloride binds with N-terminal amino acids to form dansyl polypeptides which on hydrolysis yield N-terminal dansyl amino acids. The number of dansyl amino acids produced is equal to the number of polypeptide chains in a protein.
Breakdown of the polypeptide into fragments
Polypeptides are broken down into smaller peptides by a chemical and enzymatic process.
Enzymatic cleavage
The proteolytic enzymes such as trypsin, chymotrypsin, and pepsin cleave the peptide bonds. Mostly trypsin is used. Trypsin hydrolyses the peptide bonds which contain lysine or arginine on the carbonyl side of peptide linkage.
Chemical cleavage
The chemical cyanogen bromide is commonly used to break polypeptides into smaller fragments. CNBr hydrolyses the peptide bonds which methionine on the carbonyl side.
3. Determination of amino acid sequence
Polypeptides are used for the determination of the sequence of amino acids. There are many reagents used to determine the sequence of amino acids.
Sanger’s reagent
The reagent used in sanger’s technique is 1-fluro 2, 4 dinitrobenzene. FDNB binds with the N-terminal amino acid to form a dinitro phenyl derivative of the peptide. This yields DNP amino acids which are released from the rest of the peptide chain. The DNP-amino acids can be determined by the chromatography technique.
Sanger technique has limited use as the peptide is breakdown into amino acids. The other technique used is Erdmann degradation.
Edman’s Reagent
The reagent used in edman’s technique is phenyl isothiocyanate. It reacts with the N-terminal amino acid to form a phenyl thiocarbonyl derivative. When treated with mild acid, phenyl thyohydantoin amino acid is formed. This can be identified by chromatography. The advantage of Edman’s reagent is that peptides degraded sequentially release N-terminal amino acids which can be identified. This is due to the whole peptide is not hydrolyzed.
Sequenator
Sequenator is an automatic machine used to determine the sequence of amino acids in a polypeptide. This machine is based on the principle of edman’s degradation. Amino acids are determined from N-terminal. Sequenator takes about 2 hours to determine the sequence of each amino acid.